Drug-Drug Interaction Simulation Software
DDI Simulator

Physiologically based pharmacokinetic (PBPK) model is used to quantitatively and accurately predict changes in plasma concentrations over time.
This makes it possible to evaluate drug interactions and optimize the dosage and interval of drug administration to minimize the effects of interactions.
What is DDI Simulator?
- Based on the given pharmacokinetic parameters, the plasma concentration of the substrate drug can be quantitatively predicted using a mathematical model to determine the risk of drug interaction, a necessary step in the drug development process.
- Software was developed based on the results of the Drug Interaction Database Project of the Human and Animal Bridging (HAB) Research Organization, a non-profit organization in Japan.
- New functions and data are added under the supervision of Professor Kazuya Maeda of Kitasato University School of Pharmacy and Professor Kiyomi Ito of Musashino University Faculty of Pharmacy.
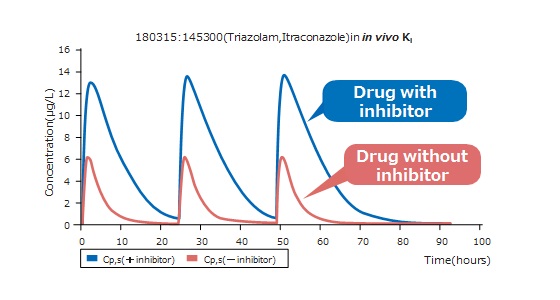
| Point1 | Proprietary in vivo Ki database |
|---|
| Point2 | Predicting drug interactions via hepatic OATPs |
|---|
| Point3 | Compliant with the Ministry of Health, Labour and Welfare's Drug Interaction Guidelines |
|---|
Product Overview
Includes a rich assortment of physiologically based pharmacokinetic (PBPK) models.
Features
Feature1 Implementing physiologically based pharmacokinetic (PBPK) models
Quantitative prediction of DDI mediated via metabolic enzyme inhibition/induction and transporter inhibition.
Feature2 Metabolic enzyme inhibition (competitive inhibition and MBI) and induction model
Predict drug interactions via competitive inhibition, MBI and induction of metabolic enzymes.
Feature3 Hepatic OATPs Inhibition Model
Predict drug interactions mediated via liver-specific OATPs.
Feature4 Drug PK Database
Representative substrates, inhibitors and inducers of major metabolic enzymes are already registered and ready to use.
Feature5 Simultaneous inhibition and induction of multiple CYP isoforms
Drug interactions can be predicted by taking into account the simultaneous inhibition and induction of multiple CYP isoforms.
Feature6 Drug Interactions Based on Inhibition & Induction of Gut Metabolism
Allows highly accurate prediction of drug interactions by taking into account the rate of change in gut availability (Fg).
Feature7 Optimizing the dosing schedule to minimize the effects of drug interactions
Dosing schedules that are less susceptible to drug interactions can be considered.
Feature8 Transporter Inhibition Model
Predicts drug interactions via transporter inhibition.
Parameter Fitting Tool
This is a tool for calculating a series of parameters that describe the kinetics of each drug in the body. A variety of available models allow for easy calculations in a few simple steps.
Available Models
- 1-compartment (no peripheral, no lag time)
- 1-compartment (no peripheral, with lag time)
- 2-compartment (with peripheral, no lag time)
- 2-compartment (with peripheral, with lag time)
- PBPK (no peripheral, no lag time)
- PBPK (no peripheral, with lag time)
- PBPK (with peripheral, no lag time)
- PBPK (with peripheral, with lag time)
Simulation Process
Assess the risk of drug interactions in just a few simple steps.
- STEP1 Import parameter fitting results (edit pharmacokinetic parameters)
- STEP2 Select a model
- STEP3 Choose drug combinations
- STEP4 Specify a dosing regimen
- STEP5 Run simulation
- STEP6 Display results
Scientific Publications
Here are some of the papers that have been published about DDI Simulator.
Mano Y, Sugiyama Y, Ito K.,
Click here for details
Use of a Physiologically Based Pharmacokinetic Model for Quantitative Prediction of Drug-Drug Interactions via CYP3A4 and Estimation of the Intestinal Availability of CYP3A4 Substrates. J Pharm Sci. 104(9):3183-93(2015)
Operating Environment
| OS | Windows 10(64bit), Windows11(64bit) |
|---|---|
| CPU | Core i5 6000 or higher |
| RAM | 4GB or more |
| Hard disk space | 1.0GB or more |
News
-
2019.07.18
- DDI Simulator Ver 2.6 released.
